Annual Report for EPSRC High End Computing Consortia 2016/17
HEC Consortium
HECBioSim (The UK High-End Computing Consortium for Biomolecular Simulation)
Consortia Chair
Adrian Mulholland (School of Chemistry, University of Bristol)
Management Group
| Adrian Mulholland (Chair) | Professor of Chemistry | University of Bristol |
| Phil Biggin | Professor of Computational Biochemistry | University of Oxford |
| Jonathan Essex | Professor of Chemistry | University of Southampton |
| Francesco Gervasio | Professor of Chemistry and Structural and Molecular Biology | UCL |
| Sarah Harris | Lecturer in Biological Physics | University of Leeds |
| Richard Henchman | Senior Lecturer in Chemistry | University of Manchester |
| David Huggins | MRC Fellow | University of Cambridge |
| Syma Khalid | Professor in Chemistry | University of Southampton |
| Charles Laughton | Associate Professor in Molecular Recognition | University of Nottingham |
| Julien Michel | Senior Lecturer | University of Edinburgh |
| Edina Rosta (ER) | Lecturer in Computational Chemistry | King’s College London |
| Mark Sansom (MS) | David Phillips Professor of Molecular Biophysics | University of Oxford |
Summary
max 2 pages
Background
HECBioSim, the UK HEC Biomolecular Simulation Consortium, was established in March 2013, and works closely with and complements CCP-BioSim (the UK Collaborative Computational Project for Biomolecular Simulation at the Life Sciences Interface). Most of the members of the Consortium are experienced users of high end computing. Biomolecular simulations are now making significant contributions to a wide variety of problems in drug design and development, biocatalysis, bio and nano-technology, chemical biology and medicine. The UK has a strong and growing community in this field, recognized by the establishment in 2011 by the EPSRC of CCP-BioSim (ccpbiosim.ac.uk), and its renewal in 2015.
There is a clear, growing and demonstrable need for HEC in this field. Members of the Consortium have, for example, served on the HECToR and ARCHER Resource Allocation Panel. The Consortium welcomes members across the whole biomolecular sciences community, and we are currently (and will remain) open to new members (unlike some consortia). Since establishing the Consortium, several new members (FLG, DH, ER) have joined the Management Group. Many of the projects awarded ARCHER time under the Consortium do not involve CCP-BioSim or HECBioSim Management Group members, demonstrating the openness of HECBioSim and its support of the biomolecular simulation community in the UK.
A number of other researchers have joined and it is our expectation that other researchers will join HECBioSim in the future. We actively engage with structural and chemical biologists and industrial researchers. We foster interactions between computational, experimental and industrial scientists (see the example case studies; members of the Consortium have excellent links with many pharmaceutical, chemical and biotechnology companies). Details of HECBioSim are available via our webpages at: http://www.hecbiosim.ac.uk.
Applications are made through the website http://www.hecbiosim.ac.uk and reviewed at one of the series of regular panel meetings.
A list of successful HECtime allocations on ARCHER is available at:http://www.hecbiosim.ac.uk/applications/successfulprojects.
All proposals are of course subject to scientific and technical review, but we have the philosophy of supporting the best science to deliver the highest impact, rather than focusing on supporting development of a couple of codes. An allocation panel (with changing membership) meets twice yearly to judge proposals and requests for AUs; projects are assessed competitively: any groups in the UK can apply. All submitted proposals receive constructive feedback from the allocation panel.
The HECBioSim website also provides forums for the biomolecular simulation community, a wiki hosting useful how-tos and user guides, and software downloads (currently FESetup and Longbow). Our lead software development project between Nottingham (Charlie Laughton and Gareth Shannon) and Daresbury (James Gebbie), we have developed a remote job submission tool, ‘Longbow’ (see below). The tool is designed to reproduce the look and feel of local MD packages, but to stage data and submit jobs to a large HPC resource, such as ARCHER, in a manner invisible to the user.
Workshops and New Opportunities
Workshops are listed below and can be found on the HECBioSim Website
We work closely with CCP-BioSim, for example in organizing training workshops and meetings. Our activities are outlined at www.hecbiosim.ac.uk Examples of forthcoming meetings include:
Computational Molecular Science 2017 - on Sunday 19 March 2017
17th European Seminar on Computational Methods in Quantum Chemistry - on Tuesday 11 July 2017
Frontiers of Biomolecular Simulation Southampton, September 2017
Issues and Problems
Details of SLA activities are given in the SLA report. Dr. James Gebbie provides support to HECBioSIm through the SLA. He has been very helpful in the construction of the HECBioSim webpages (hecbiosim.ac.uk). James also worked with Dr. Gareth Shannon on the development of Longbow (See above).
In the past year, members of the Consortium faced some queuing problems; close to the end of the first allocation period in particular, there were long queuing times, which led to problems in completing jobs and using allocated time. Throughput has been a real problem in the past few months.
Membership
Please provide a full list of existing members and their institutions, highlighting any new members that have joined the consortium during the reporting period. If available please provide information on the number of distinct users that have accessed ARCHER via the Consortium during this reporting period.
Below is a list of PIs with HECBioSim projects in the current reporting period:
| Agnes Noy | University of York | new to this reporting period |
| Alessandro Pandini | Brunel University London | |
| Arianna Fornili | Queen Mary University of London | |
| Cait MacPhee | University of Edinbrurgh | new to this reporting period |
| Charlie Laughton | University of Nottingham | |
| Clare-Louise Towse | University of Bradford | new to this reporting period |
| D Flemming Hansen | University College London | new to this reporting period |
| David Huggins | Aston University | |
| Edina Rosta | King's College London | |
| Francesco Gervasio | University College London | |
| Ian Collinson | University of Bristol | new to this reporting period |
| Irina Tikhonova | Queen's University Belfast | |
| Jiayun Pang | University of Greenwich | |
| Jonathan Doye | University of Oxford | |
| Julien Michel | University of Edinburgh | |
| Mario Orsi | UWE Bristol | |
| Michele Vendruscolo | University of Cambridge | |
| Michelle Sahai | University of Roehampton | |
| Philip Biggin | University of Oxford | |
| Richard Sessions | University of Bristol | |
| Stephen Euston | Heriot-Watt University | |
| Syma Khalid | University of Southampton |
PIs in HECBioSIm outside of current allocation period
| Peter Bond | University of Cambridge |
| Richard Bryce | University of Manchester |
| Juan Antonio Bueren-Calabuig | University of Edinburgh |
| Christo Christov | Northumbria University |
| Anna K Croft | University of Nottingham |
| Jonathan Essex | University of Southampton |
| Robert Glen | University of Cambridge |
| Sarah A Harris | University of Leeds |
| Jonathan Hirst | University of Nottingham |
| Douglas Houston | University of Edinburgh |
| Dmitry Nerukh | Aston University |
| Andrei Pisliakov | University of Dundee |
| Mark Sansom | University of Oxford |
| Marieke Schor | University of Edinburgh |
| Gareth Shannon | University of Nottingham |
| Tatyana Karabencheva-Christova | Northumbria University |
There are currently 164 members of HECBioSim (i.e. users of HECBioSIm ARCHER time), including the above PIs. 20 new users have been added since January 2016.
World Class and World Leading Scientific Output
ARCHER should enable high quality and world-leading science to be delivered. This should generate high impact outputs and outcomes that increase the UK’s position in world science.
- If all the publications relating to the work of the Consortium for this reporting period have been added to ResearchFish / will be added to ResearchFish by the end of the ResearchFish reporting exercise, please indicate this below.
- If submission of a full list of publications to the Consortium record/s in ResearchFish has not been possible for this reporting period please provide a list of publications that have resulted from work performed on ARCHER by the Consortium during this reporting period (this can be included as a separate attachment).
- For the reporting period please provide a bullet pointed list of key / important research findings that has resulted from work performed on ARCHER by the Consortium. Please reference any related publications.
- For the reporting period please include a bullet pointed list of any relevant press announcements and other communications of significance to an international community.
All publications can be found in ResearchFish, and have been added by individual researchers. A selection of illustrative highlights are provided below:
Julien Michel (University of Edinburgh)
New molecular simulation methods have enabled for the description of first time a complete the interactions of several drug-like small molecules with an intrinsically disordered region of the oncoprotein MDM2, as well as the effect of post-translational modifications on the dynamics of this protein. The results obtained suggest new medicinal chemistry strategies to achieve potent and selective inhibition of MDM2 for cancer therapies.
Publication/s:
Elucidation of Ligand-Dependent Modulation of Disorder-Order Transitions in the Oncoprotein MDM2 http://journals.plos.org/ploscompbiol/article?id=10.1371/journal.pcbi.1004282
Impact of Ser17 Phosphorylation on the Conformational Dynamics of the Oncoprotein MDM2 http://pubs.acs.org/doi/abs/10.1021/acs.biochem.6b00127
An outreach document is being prepared by EPCC to showcase a lab project on isoform selective inhibition that has benefited from both HECBioSim and EPCC ARCHER allocations.
Syma Khalid (Chemistry, Southampton)
Publication/s:
OmpA: A Flexible Clamp for Bacterial Cell Wall Attachment Samsudin F, Ortiz-Suarez ML, Piggot TJ, Bond PJ, Khalid S (2016). “OmpA: a Flexible Clamp for Bacterial Cell Wall Attachment”, Structure, 24(12):2227-2235
With Singapore National Center for Biotechnology.
Sarah Harris (Physics, Leeds)
In collaboration with the experimental biochemistry group lead by Radford at Leeds, Sarah Harris used ARCHER time to show how chaperones help outer membrane proteins to fold.
Publication/s:
Schiffrin B, Calabrese AN, Devine PWA, Harris SA, Ashcroft AE, Brockwell DJ, Radford SE “Skp is a multivalent chaperone of outer-membrane proteins” Nature Structural and Molecular Biology 23 786-793, 2016. DOI:10.1038/nsmb.3266
Michelle Sahai (Department of Life Sciences, Roehampton)
Publication/s:
Combined in vitro and in silico approaches to the assessment of stimulant properties of novel psychoactive substances - The case of the benzofuran 5-MAPB. Progress in Neuro-Psychopharmacology and Biological Psychiatry 75, Pages 1–9 (2017). https://www.ncbi.nlm.nih.gov/pubmed/27890676
Phil Biggin (Biochemistry, Oxford)
Outputs that have used HECBioSim time:
1. Steered Molecular Dynamics Simulations Predict Conformational Stability of Glutamate Receptors. Musgaard M, Biggin PC. J Chem Inf Model. 2016 Sep 26;56(9):1787-97. doi: 10.1021/acs.jcim.6b00297.
2. Kainate receptor pore-forming and auxiliary subunits regulate channel block by a novel mechanism. Brown PM, Aurousseau MR, Musgaard M, Biggin PC, Bowie D. J Physiol. 2016 Apr 1;594(7):1821-40. doi: 10.1113/JP271690.
3. Role of an Absolutely Conserved Tryptophan Pair in the Extracellular Domain of Cys-Loop Receptors. Braun N, Lynagh T, Yu R, Biggin PC, Pless SA. ACS Chem Neurosci. 2016 Mar 16;7(3):339-48. doi: 10.1021/acschemneuro.5b00298.
4. Distinct Structural Pathways Coordinate the Activation of AMPA Receptor-Auxiliary Subunit Complexes. Dawe GB, Musgaard M, Aurousseau MR, Nayeem N, Green T, Biggin PC, Bowie D. Neuron. 2016 Mar 16;89(6):1264-76. doi: 10.1016/j.neuron.2016.01.038.
Covered by 7 news outlets:
I. Health Canal (http://www.healthcanal.com/brain-nerves/70646-what-makes-the-brain-tick-so-fast.html)
II. Health Medicine Network (http://healthmedicinet.com/i/scientists-reveal-how-the-brain-processes-information-with-lightning-speed/)
III. News Medical (http://www.news-medical.net/news/20160227/Scientists-reveal-how-the-brain-processes-information-with-lightning-speed.aspx)
IV. Wired (http://www.wired.co.uk/article/what-we-learned-about-the-brain-this-week-3)
V. Alpha Galileo (http://www.alphagalileo.org/ViewItem.aspx?ItemId=161473&CultureCode=en)
VI. Science Daily (https://www.sciencedaily.com/releases/2016/02/160225140254.htm)
VII. Eureka Alert (https://www.eurekalert.org/pub_releases/2016-02/mu-wmt022516.php)
Recommended by F1000 Prime:
http://f1000.com/prime/726180600.
5. Accurate calculation of the absolute free energy of binding for drug molecules. Aldeghi M, Heifetz A, Bodkin MJ, Knapp S, Biggin PC. Chem Sci. 2016 Jan 14;7(1):207-218.
Arianna Fornili (Biological and Chemical Sciences, Queen Mary University of London)
Publication/s:
Fornili, E. Rostkova, F. Fraternali, M. Pfuhl: Effect of RlC N-Terminal Tails on the Structure and Dynamics of Cardiac Myosin. Biophysical J. 110 (2016) 297A.
More in preparation.
Dmitry Nerukh (Engineering and Applied Science, Aston University)
Important research findings: Distribution of ions inside a viral capsids influences the stability of the capsid.
Edina Rosta (Computational Chemistry, Kings College London)
Highlighted publication:
We have a joint experimental paper with the Vertessy group in JACS where we identified a novel arginine finger residue in dUTPase enzymes, and described the mechanism of action for this residue using crystallographic data, biochemical experiments, MD and QM/MM simulations thanks to HECBioSim. http://pubs.acs.org/doi/abs/10.1021/jacs.6b09012
Press release:
Our joint paper with Jeremy Baumberg's group in Cambridge was published in Nature. http://www3.imperial.ac.uk/newsandeventspggrp/imperialcollege/newssummary/news_13-6-2016-16-52-4
Greater Scientific Productivity
As well as speed increases, the optimisation of codes for the ARCHER machine will enable problems to be solved in less time using fewer compute resources.
For the reporting period please provide a brief update on the progress of software development activities associated with the Consortium and the impact this has had on Consortium members and the broader research community.
Our lead software development project between Nottingham (Charlie Laughton and Gareth Shannon) and Daresbury (James Gebbie), we have developed a remote job submission tool, ‘Longbow’ (see below). The tool is designed to reproduce the look and feel of local MD packages, but to stage data and submit jobs to a large HPC resource such as ARCHER in a manner invisible to the user. An open beta version was released unrestricted to the community in March 2015 (http://www.hecbiosim.ac.uk/longbow and via PyPI https://pypi.python.org ) with ~100 downloads. Functionality includes native support for biosimulation packages AMBER, CHARMM, GROMACS, LAMMPS and NAMD, native support for jobs on ARCHER, native support for jobs running on PBS and LSF schedulers, and support for three different job types (single, ensembles and multiple jobs).
The software is written both as an application for users and an API for developers. CCP-EM have integrated Longbow into their developmental GUI, and shortly FESetup will ship with native support for job submission using Longbow. We are currently in talks with ClusterVision with respect to them distributing Longbow to their user base as a user friendly way for novices to interact with their systems.
Longbow
Longbow has been growing in popularity both amongst researchers within the biosimulation field and those in other fields. In this reporting period there have been five new releases of Longbow delivering a plethora of user requested features and bug fixes. Some of the key developments are summarised below:
- Implemented a Recovery mode – Should the event happen that Longbow crashes or the system in which Longbow is controlling jobs from powers down. The user can now reconnect to the crashed session and carry on as if nothing happened.
- Sub-Queuing – More and more system administrators are setting limits not just on the number of simulations that can run, but also on the number of jobs that can go into the queue. Longbow can now automatically detect this and implement its own queue feeding jobs into the system queue as slots open up.
- Dis-connectible/Re-connectible Longbow sessions – A user can now launch Longbow to fire off all jobs and then disconnect, at a later date the user can re-establish the connection and download all results (no need for persistent connections anymore).
- Ability to include scripts in the Longbow generated submit files.
- Numerous stability, performance and bug fixes.
Longbow continues to be the submission engine that is used under the hood of the CCP-EM toolkit FLEX-EM. There are several other interesting projects that are making use of Longbow.
- A project by the energy efficient computing group (Hartree Center) are currently integrating Longbow with CK, a crowd sourced compilation optimisation tool aimed at finding efficient compilation configuration.
- A project by the Applications performance engineering group (Hartree Center) are currently integrating longbow into Melody, a runtime optimisation tool aimed at optimising runtime configuration of simulations.
- A project by the computational biology group (Hartree Center) an automated setup, launch and analysis platform for MD on protein-membrane simulations.
Metrics: Downloads (HECBioSim) 985 Downloads (PyPi) 3456
Increasing the UK’s CSE Skills Base (including graduate and post doctorate training and support)
This builds on the skills sets of trained people in HPC, both in terms of capacity and raising the overall skill level available to the sector.
- For the reporting period please provide information on the number of PhDs and Post-Docs that have been trained in the use of ARCHER as a result of work relating to the Consortium.
- For the reporting period please provide a bullet pointed list of training activities undertaken by the Consortium, providing information on the target audience and level of attendance.
125 PhD and PDRAs have been trained and have used ARCHER through HECBioSim. In the past 6 months alone that has included:
PhD students:
| Marc Dämgen | University of Oxford |
| Laura Domicevica | University of Oxford |
| Matteo Aldeghi | University of Oxford |
| Shaima Hashem | Queen Mary University of London |
| Ruth Dingle | University College London |
| Lucas Siemons | University College London |
| Elvira Tarasova | Aston University |
| Vladimiras Oleinikovas | University College London |
| Havva Yalinca | University College London |
| Daniel Moore | Queen’s University Belfast |
| Aaron Maguire | Queen’s University Belfast |
| Maxime Tortora | University of Oxford |
| Michail Palaiokostas | UWE Bristol |
| Wei Ding | UWE Bristol |
| Ganesh Shahane | UWE Bristol |
| Pin-Chia Hsu | University of Southampton |
| Damien Jefferies | University of Southampton |
PDRA:
| Maria Musgaard | University of Oxford |
| Teresa Paramo | University of Oxford |
| Marieke Schor | University of Edinbrurgh |
| Antonia Mey | University of Edinbrurgh |
| Ioanna Styliari | University of Nottingham |
| Ivan Korotkin | Aston University |
| Silvia Gomez Coca | King’s College London |
| Giorgio Saladino | University College London |
| Federico Comitani | University College London |
| Robin Corey | University of Bristol |
| Jordi Juarez-Jimenez | University of Edinburgh |
| Predrag Kukic | University of Cambridge |
| Giulia Tomba | University of Cambridge |
| Deborah Shoemark | University of Bristol |
| Georgios Dalkas | Heriot-Watt University |
| Robin Westacott | Heriot-Watt University |
| Firdaus Samsudin | University of Southampton |
| Agnes Noy | University of Leeds (now EPSRC fellow at York) |
Please see below details of the training week held in June 2016 at the University of Bristol. 170 delegates attending the training which ran over 5 days. There is course content available on the CCPBioSim Workshop Page.
CCPBioSim Training Week - Day 5: QM/MM enzyme reaction modelling - on Friday 10 June 2016
CCPBioSim Training Week - Day 4: Monte Carlo Methods for Biomodelling - on Thursday 09 June 2016
CCPBioSim Training Week - Day 3: Python for Biomodellers and FESetup - on Wednesday 08 June 2016
CCPBioSim Training Week - Day 2: Running and analysing MD simulations - on Tuesday 07 June 2016
CCPBioSim Training Week - Day 1: Enlighten: Tools for enzyme-ligand modelling - on Monday 06 June 2016
Past Workshops and Conferences:
AMOEBA advanced potential energies workshop - on Friday 09 December 2016
Intel Training Workshop (Parallel programming and optimisation for Intel architecture) - on Wednesday 30 November 2016
3rd Workshop on High-Throughput Molecular Dynamics (HTMD) 2016 - on Thursday 10 November 2016
Free Energy Calculation and Molecular Kinetics Workshop - on Tuesday 13 September 2016
Going Large: tools to simplify running and analysing large-scale MD simulations on HPC resources - HECBioSim - on Wednesday 16 December 2015
This 1 day workshop dealt with Longbow, a Python tool created by HECBioSim consortium that allows use of molecular dynamics packages (AMBER, GROMACS, LAMMPS, NAMD) with ease from the comfort of the desktop, and pyPcazip, a flexible Python-based package for the analysis of large molecular dynamics trajectory data sets.
Richard Henchman (Chemistry, Manchester)
I lectured on the CCP5 Summer School in 2015 in Manchester and 2016 in Lancaster for the Advanced Topic Biomolecular Simulation drawing on CCPBiosim training materials.
Sarah Harris (Physics, Leeds) and Agnes Noy
Agnes Noy was trained to use ARCHER independently which helped her to obtain an Early-career EPSRC fellowship. The fellowship grant follows the study of supercoiling DNA by modelling (further funding) and introduces a new member to the community (grant is EPSRC (EP/N027639/1)).
Increased Impact and Collaboration with Industry
ARCHER does not operate in isolation and the ‘impact’ of ARCHER’s science is converted to economic growth through the interfaces with business and industry. In order to capture the impacts, which may be economic, social, environmental, scientific or political, various metrics may be utilised.
- For the reporting period please provide where possible information on Consortium projects that have been performed in collaboration with industry, this should include:
1. Details of the companies involved.
2.Information on the part ARCHER and the Consortium played.
3. A statement on the impact that the work has / is making.
4. If relevant, details of any in kind or cash contributions that have been associated with this work. - For the reporting period include a list of Consortium publications that have industrial co-authorship.
- For the reporting period please provide details of the any other activities involving industrial participation e.g. activities involving any Industrial Advisory panels, attendance / participation in workshops and Consortium based activities.
Examples of industrial engagement through HECBioSim include:
Syma Khalid (Computational Biophysics, Southampton)
Collaboration with Siewert-Jan Marrink (Netherlands) and Wonpil Im(USA) - paper is being written at the moment - also we have developed and tested the following computational tool as part of this project: http://www.charmm-gui.org/?doc=input/membrane
We have signed an NDA with a company developing antibiotics (Auspherix) based on work published in Samsudin et al, Structure, 2016. The company provide us with experimental data and crucially the structures of their potential drug molecules, and we work out how they interact with the bacterial membrane.
Press release - https://www.sciencedaily.com/releases/2016/11/161121094122.htm
ARCHER time we got from HECBioSim was essential for this project. These are large simulations that are very computationally demanding.
Julien Michel (Chemistry, Edinburgh)
Industrial collaborations that have benefited from HECBioSim
UCB was a project partner of EPSRC-funded research in the Michel lab on modelling binding of ligands to flexible proteins. HECBioSim supported the research via allocation of computing time on ARCHER. The work has been published.
Impact of Ser17 phosphorylation on the conformational dynamics of the oncoprotein MDM2 Bueren-Calabuig, J. A. ; Michel, J. Biochemistry, 55 (17), 2500-2509, 2016
Elucidation of Ligand-Dependent Modulation of Disorder-Order Transitions in the Oncoprotein MDM2 Bueren-Calabuig, J. A. ; Michel, J. PLoS Comput. Biol. , 11(6): e1004282, 2015
International activities: I represented HECBioSim at the MolSSI conceptualisation workshop in Houston (October 2016).
Arianna Fornili (School of Biological and Chemical Sciences, Queen Mary University of London)
This project is done in collaboration with experimental (NMR and SAXS) partners at King’s College London (Dr. Mark Pfuhl and Dr. Elena Rostkova), with the final aim of providing a complete molecular model of how muscle contraction is regulated in the heart.
Francesco Gervasio (Chemistry, UCL)
A new collaboration with UCB on developing new approaches to sample cryptic binding sites of pharmaceutical interest was started. UCB co-sponsored a BBSRC-CASE PhD studentships. The code development and its application to interesting targets was made possible by the access to Archer.
Phil Biggin (Biochemistry, Oxford)
The following has industrial partners as co-authors:
Accurate calculation of the absolute free energy of binding for drug molecules, Aldeghi M, Heifetz A, Bodkin MJ, Knapp S, Biggin PC. Chem. Sci., 2016,7, 207-218 doi: 10.1039/C5SC02678D. The GLAS scoring module for gromacs has yet to be implemented but that would also constitute industrial activity.
Richard Henchman (Chemistry, Manchester)
I have one publication with an industrial co-author (Evotec), which Julien Michel is part of as well.
Assessment of hydration thermodynamics at protein interfaces with grid cell theory, G. Gerogiokas, M. W. Y. Southey, M. P. Mazanetz, A. Heifetz, M. Bodkin, R. J. Law, R. H. Henchman and J. Michel, J. Phys. Chem. B, 2016, 120, 10442–10452.
Strengthening of UK's International Position
The impacts of ARCHER’s science extend beyond national borders and most science is delivered through partnerships on a national or international level.
- For the reporting period please provide a bullet pointed list of projects that have involved international collaboration. For each example please provide a brief summary of the part that ARCHER and the Consortium have played.
- For the reporting period please provide a list of consortium publications with international co-authorship.
- For the reporting period please detail any other international activities that the Consortium might be involved in (workshops, EU projects etc.).
Examples of HECBioSim international engagement include:
Michelle Sahai (Department of Life Sciences, Roehampton)
Combined in vitro and in silico approaches to the assessment of stimulant properties of novel psychoactive substances - The case of the benzofuran 5-MAPB. Sahai MA, Davidson C, Khelashvili G, Barrese V, Dutta N, Weinstein H, Opacka-Juffry J. Prog Neuropsychopharmacol Biol Psychiatry. (2016) 75:1-9. doi: 10.1016/j.pnpbp.2016.11.004.
co-authors affiliations include:
Department of Physiology and Biophysics, Weill Cornell Medical College of Cornell University (WCMC), New York, NY, 10065, USA and HRH Prince Alwaleed Bin Talal Bin Abdulaziz Alsaud Institute of Computational Biomedicine, Weill Cornell Medical College of Cornell University, New York, NY, 10065, USA.
Author contribution:
CD, MAS, HW and JOJ were responsible for the study concept and design. CD and VB collected and interpreted the voltammetry data. JOJ and ND conducted the ligand binding experiments and ND analysed the data. MAS (ARCHER user) and GK performed the molecular modelling studies and interpreted the findings. CD, MAS, GK and JOJ drafted the manuscript. All authors critically reviewed the content and approved the final version for publication.
Dmitry Nerukh (Engineering and Applied Science, Aston University)
Collaborations:
Prof. Makoto Taiji group (K-computer, MDGRAPE), Laboratory for Computational Molecular Design, Computational Biology Research Core, RIKEN Quantitative Biology Center (QBiC), Kobe, Japan
Prof. Reza Khayat group, City College of New York, United States
Prof. Artyom Yurov group, Immanuel Kant Baltic Federal University, Russian Federation
Prof. Nikolay Mchedlov-Petrosyan group, V.N. Karazin Kharkiv National University, Ukraine
Prof. Natalya Vaysfeld group, Odessa I.I.Mechnikov National University, Ukraine
International activities:
International Workshop - Engineering bacteriophages for treating antimicrobial resistance using computational models. Dec. 2016, Aston University, UK
Dmitry Nerukh group at Aston University collaborates with the group of Prof. Makoto Taiji (the deputy director of RIKEN Quantitative Biology Institute). Prof. Taiji group is the author and implementer of the fastest machine in the world for molecular dynamics simulations, MDGRAPE, as well as part of the team designing the K-computer and working on it (several times the fastest computer in the world). We are currently preparing a joint manuscript for publication where the results obtained on ARCHER will be used alongside with the results obtained on MDGRAPE-4.
Julien Michel (Chemistry, Edinburgh)
Free Energy Reproducibility Project
Free energy reproducibility project between the Michel, Mobley, Roux groups and STFC.
This makes use of FESetup which is CCPBioSim, but calculations have been executed on various HPC platforms, Hannes can clarify whether any of the systems used is within the remit of HECBioSim.
Sarah Harris (Physics, Leeds), Charlie Laughton (Pharmacy, Nottingham) and Agnes Noy (Physics, York)
In an international collaboration with the Institute for Research in Biomedicine in Barcelona, Noy, Harris and Laughton used ARCHER time obtained through the HEBioSim consortium to perform multiple 100ns molecular dynamics (MD) simulations of DNA minicircles containing ~100 base pairs as part of the validation suite for the new BSC1 nucleic acid forcefield, see:
Ivani I., Dans P. D., Noy A., Perez A., Faustino I., Hospital A., Walther J., Andrio P., Goni R., Portella G., Battistini F.,Gelpi J. L. Gonzalez C., Vendruscolo M., Laughton C. A., Harris S. A. Case D. A. & Orozco M. “Parmbsc1: a refined force field for DNA simulations” Nat. Methods 13, 55, 2015, doi:10.1038/nmeth.3658
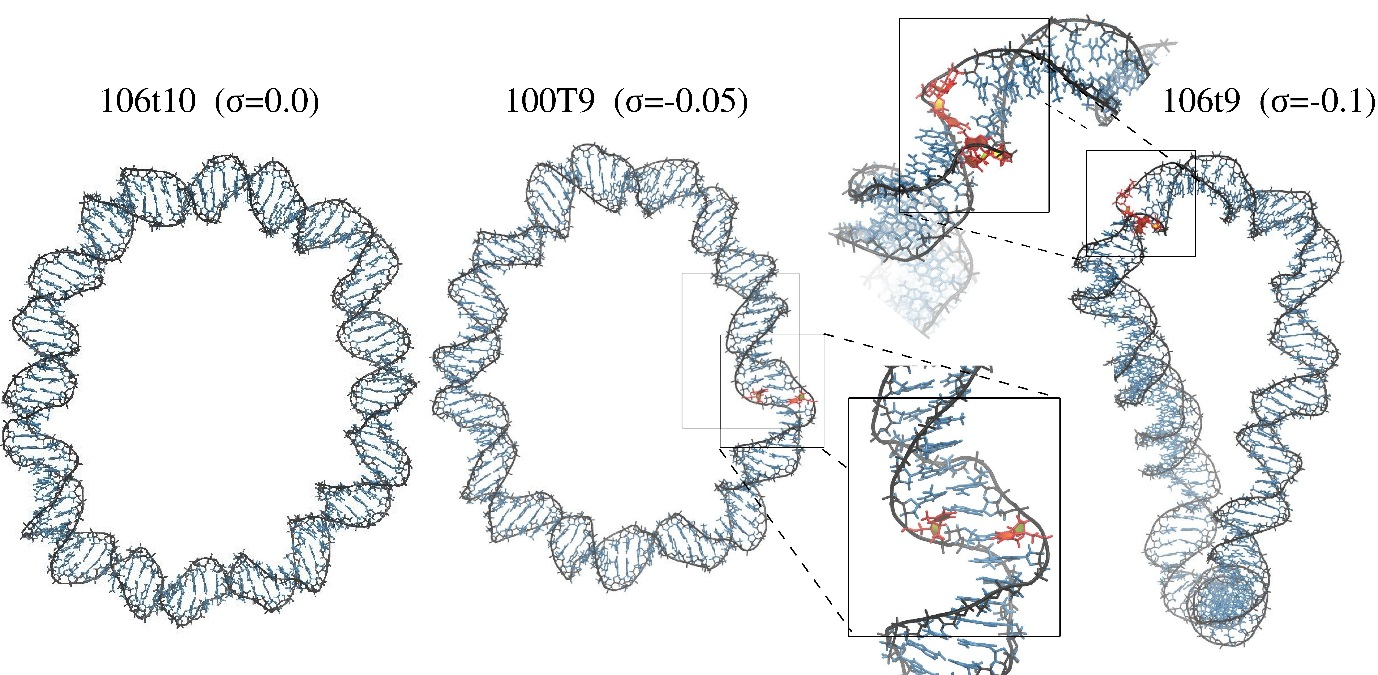
MD simulations of minicircles. Molecular structures showing the final frame of the minicircles MD simulations.
The secondary structure of the relaxed loop with 106 bp and 10 helical turns (106t10) remains intact, while the 2 negatively supercoiled circles denature.
The 100 bp circle with 9 tuns (100t9) presents 2 adjacent pyrimidine base-flippings towards major groove,
and the 106 bp circle with 9 turns (106t9) denature over multiple consecutive base pairs.
Jon Essex (Chemistry, Southampton)
Development and testing of hybrid coarse-grain/atomistic model of membrane systems. ARCHER and the consortium were instrumental in providing the computing time necessary to complete this work. The work was performed by a research fellow in my group, who has subsequently taken up an academic post in Sweden, where he is continuing to develop this methodology.
Publication: All-atom/coarse-grained hybrid predictions of distributioncoefficients in SAMPL5 Samuel Genheden1, Jonathan W. Essex, J Comput Aided Mol Des (2016) 30:969–976 DOI 10.1007/s10822-016-9926-z
IP and other industrial engagement, or translation, which has benefited from HECBioSim international collaborations
Collaboration Dr Samuel Genheden, University of Gothenburg – see above
International activities (e.g. workshops) that have benefited from HECBioSim
The work performed with Sam resulted in an successful eCSE application for development work on the LAMMPS software. This is an international simulation package run out of Sandia labs in the US. Through this eCSE application we were able to update the rotational integrator and improve the load balancing for our hybrid simulation methodology. These developments have been incorporated in the latest release versions of the code. An ARCHER training webinar resulted from this work.
Training activities
see above – ARCHER training webinar on our developments within LAMMPS
Software development
see above – code modifications in LAMMPS which either have been, or will be, in the release version of the code
Advantages you see of the consortium (e.g. could be ability to pursue project quickly; develop collaborations…) - resources to run the sort of large-scale calculations we need
Michelle Sahai (Department of life Sciences, Roehampton)
The publication I added on ResearchFish has international co-authors.
Combined in vitro and in silico approaches to the assessment of stimulant properties of novel psychoactive substances - The case of the benzofuran 5-MAPB. Sahai MA, Davidson C, Khelashvili G, Barrese V, Dutta N, Weinstein H, Opacka-Juffry J. Prog Neuropsychopharmacol Biol Psychiatry. (2016) 75:1-9. doi: 10.1016/j.pnpbp.2016.11.004.
co-authors affiliations include:
Department of Physiology and Biophysics, Weill Cornell Medical College of Cornell University (WCMC), New York, NY, 10065, USA and HRH Prince Alwaleed Bin Talal Bin Abdulaziz Alsaud Institute of Computational Biomedicine, Weill Cornell Medical College of Cornell University, New York, NY, 10065, USA.
Author contribution:
CD, MAS, HW and JOJ were responsible for the study concept and design. CD and VB collected and interpreted the voltammetry data. JOJ and ND conducted the ligand binding experiments and ND analysed the data. MAS (ARCHER user) and GK performed the molecular modelling studies and interpreted the findings. CD, MAS, GK and JOJ drafted the manuscript. All authors critically reviewed the content and approved the final version for publication.
Edina Rosta (Computational Chemistry, Kings College London)
International collaborations
I have several international collaborators with joint computational/experimental projects. For their success the HECBioSim computer time was essential:
Prof. Beata Vertessy, Budapest Technical University, Hungary
JACS 2016 138 (45), 15035-15045
Profs. Walter Kolch, Vio Buchete and Boris Kholodenko, UCD, Ireland
Angewandte 55 (3), 983-986, 2016 (this may have been reported previously)
PLOS Computational Biology 12 (10), e1005051, 2016
J Phys Chem Letters 7 (14), 2676-2682, 2016
Jose Maria Lluch, Angels Gonzalez, Autonomous University of Barcelona, Spain
JCTC 12 (4), 2079-2090, 2016
Other international activities
Free energy and molecular kinetics workshop with Frank Noe's group (Free University of Berlin) and Vio Buchete (UCD, Dublin). Maybe this should be for training activities?
Following the workshop, Erasmus students apply to visit my group.
Phil Biggin (Biochemistry, Oxford)
1 and 3 were with colleagues from McGill (Brown PM, Aurousseau MR and Bowie) and 2 was with colleagues from Denmark (Braun and Pless)
1. Kainate receptor pore-forming and auxiliary subunits regulate channel block by a novel mechanism. Brown PM, Aurousseau MR, Musgaard M, Biggin PC, Bowie D. J Physiol. 2016 Apr 1;594(7):1821-40. doi: 10.1113/JP271690.
2. Role of an Absolutely Conserved Tryptophan Pair in the Extracellular Domain of Cys-Loop Receptors. Braun N, Lynagh T, Yu R, Biggin PC, Pless SA. ACS Chem Neurosci. 2016 Mar 16;7(3):339-48. doi: 10.1021/acschemneuro.5b00298.
3. Distinct Structural Pathways Coordinate the Activation of AMPA Receptor-Auxiliary Subunit Complexes. Dawe GB, Musgaard M, Aurousseau MR, Nayeem N, Green T, Biggin PC, Bowie D. Neuron. 2016 Mar 16;89(6):1264-76. doi: 10.1016/j.neuron.2016.01.038.
Richard Henchman (Chemistry Manchester)
I have an international collaboration with Professor Franke Grater at the University of Heidelberg on entropy theory for biomolecular systems.
Syma Khalid (Chemistry, Southampton)
Collaboration with Singapore National Center for Biotechnology.
Contributions to CECAM workshops
Other Highlights for the Current Reporting Period
Please provide details of any other significant highlights from the reporting period that are not captured elsewhere in the report.
Professor Adrian Mullholland organised and chaired the Computational Chemistry, Gordon Research Conference, Girona, Spain 24th – 29th July 2016.
The theme of the 2016 Computational Chemistry GRC was "Theory and Simulation Across Scales in Molecular Science”. It focused on method development and state-of-the-art applications across computational molecular science, and encouraged cross-fertilization between areas. The conference was oversubscribed, with attendees from industry and academia. The meeting received a High-Performance Rating, and was commended for the assessment that 95% of conferees rated this meeting "above average" on all evaluation areas (science, discussion, management, atmosphere and suitability).
HEC Consortia Model
Over the coming months EPSRC will be looking at the future of the HEC Consortia model and potential future funding. We would like to use this opportunity to ask the Consortia Chairs for input:
- What are the key benefits that your community have experienced through the existence of the HEC Consortia?
- What elements of the financial support provided by the HEC Consortium’s grant have worked well and what could be improved in the future?
We aim to expand the breadth of the work of the Consortium focusing on cutting-edge applications, and building collaborations with experiments and industry, to achieve maximum impact from ARCHER use. We discussed Grand Challenges at our most recent Management Group meeting. In December 2016, and have identified several to follow up as strategic priorities. We aim to tackle and support large-scale grand challenge applications in biomolecular science, in areas such as antimicrobial resistance, membrane dynamics, drug design and synthetic biology.
One specific theme with potentially high impact in drug discovery is large -scale comparative investigation of allosteric regulation in different superfamilies of proteins (e.g. PAS domain containing proteins, tyrosine kinases, etc.). The major impact will be in the identification of novel selective therapeutic molecules with limited adverse side effects.As a Consortium, we intend to develop and apply novel computational approaches for the rational design of allosteric regulators of hitherto ‘undruggable’ targets. Many of the targets emerging from large-scale genetic screening are deemed undruggable due to the difficulty of designing drug-like modulators that bind to their catalytic sites. It is increasingly clear that these targets might be effectively targeted by designing drugs that bind to protein-protein interfaces and allosteric sites. To do that, however, new rational design strategies, based on an in-depth knowledge of protein dynamics and advanced modelling and new simulation techniques are required.
Other challenging frontier areas include dynamics of motor proteins; prediction of the effects of pathogenic mutations on protein function. The accurate prediction of free energy of binding is still in its infancy and needs much further investigation. Finally, kinetics of biomolecular reactions will become the next big topic. It is clear that high end computing resource can provide the necessary power and capability to provide inroads into this vital area. This is important because it is becoming increasingly apparent that kinetics of drug binding is a key factor that the pharmaceutical sector should really be looking at in terms of a major dictator of potency.
HECBioSim is now well established, supporting work of many groups across the UK in the growing field of biomolecular simulation. We benefit from an Advisory Group containing members from industry, as well as international biomolecular simulation experts and experimental scientists. The Advisory Board was expanded and refreshed for the renewal and consists of: Dr. Nicolas Foloppe (Vernalis plc, Chair); Dr. Colin Edge (GlaxoSmithKline); Dr. Mike King (UCB Pharmaceuticals); Dr. Mike Mazanetz (Evotec AG); Dr. Garrett Morris (Crysalin Ltd.); Dr. Gary Tresadern (Johnson and Johnson Pharmaceuticals); Dr. Richard Ward (AstraZeneca); Prof. Modesto Orozco (IRB, Barcelona); Prof. Tony Watts, (Oxford, NMR); Dr. Pete Bond (A*STAR Bioinformatics Institute Singapore). We aim to foster industrial collaborations and collaborations with experimentalists (e.g. joint workshops with CCPN, Institute of Physics (Sarah Harris, Leeds Physics); see e.g. case studies. A particular theme for strategic development will be multiscale modelling, building on collaborations between several groups in the Consortium and the other CCPs. This theme reflects the inclusive and forward looking philosophy of HecBioSim, which is a community open to new ideas, keen to develop new methods, and ultimately to use HEC to drive exciting new science.
The Consortium model allows new collaborations to develop. The recent reduction in time allocated to HECBioSim has meant that we can support fewer projects. There is significantly more demand for computer time than we can accommodate through our current allocation. We intend to work with Tier 2 Centres to explore possibilities for applications, and e.g. to test emerging architectures for biomolecular simulation.
Edina Rosta (Computational Chemistry, Kings College London) comments: ”HECBioSim enables my group to perform biomolecular simulations at the high standards required for JACS, Angewandte, etc. Without this consortium I would not be able to perform the necessary calculations leading to publishable work in top journals as our local university resources are limited and the HPC systems are not well maintained”.
Regarding financial support: the administrative support provided via the consortium grant is essential to the functioning of HECBioSim. This role is currently undertaken by Simone Breckell who not only administers, allocates time to projects and arranges panel meetings but has also done a fantastic job collating the extensive information for this EPSRC report.
Note: The case studies submitted with this report have been omitted from this online document. The production case studies can be found on this website under the case studies menu heading.
